Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
U-Net was published in 2015 for spotting microscopic cells in biomedical scans, and since then, it became very popular. It created a massive impact; before U-Net, models were using a sliding window approach, and this process took a lot of time. U-Net processed the image at once, reducing training time drastically. This made real-time segmentation actually possible. Another notable impact was the number of training images; U-Net had augmentation methods that reduced the number of training images required for training a decent model. In this article, I will show you how to train a U-Net semantic segmentation model with any dataset using PyTorch.

There will be 4 main steps:
Also, I have a YouTube video about this article, you can watch it.
For training models, you need to have a GPU-supported PyTorch environment. If you don’t have a GPU, or don’t want to spend time on installations, you can directly use Kaggle or Google Colab. In the GitHub repository, there are different installation methods, including Docker installation, you can check. Now, I will show you the simplest one. You can create a new conda environment or a virtual environment.
First, let’s create a new environment:
conda create -n unet python=3.10 -y
conda activate unet
Let’s install PyTorch with pip; depending on your CUDA version, you should change the PyTorch version. If you get any error, you can read my article about how to create a GPU-supported PyTorch environment (link).
pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu124
Now, let’s clone the U-Net repository and install helper libraries:
git clone https://github.com/milesial/Pytorch-UNet.git
cd Pytorch-UNet
pip install -r requirements.txt
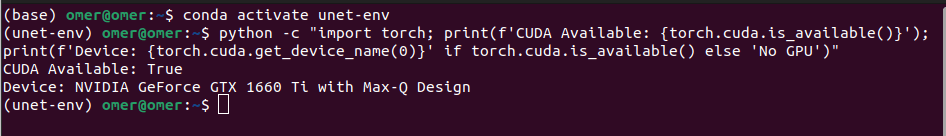
From the terminal, you can check if a GPU is available within your environment:
python -c "import torch; print(f'CUDA Available: {torch.cuda.is_available()}'); print(f'Device: {torch.cuda.get_device_name(0)}' if torch.cuda.is_available() else 'No GPU')"
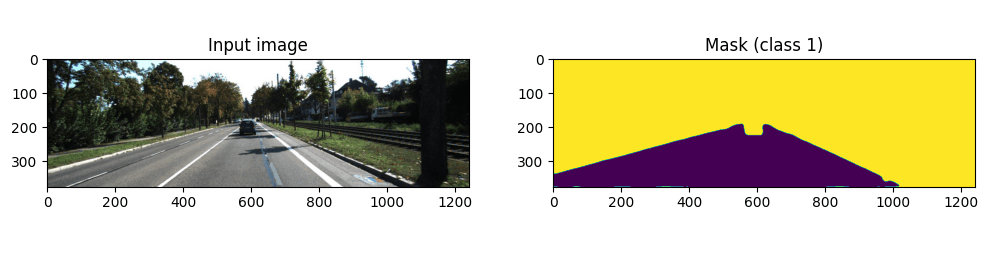
I randomly chose a dataset from Roboflow (link), you are free to choose any semantic segmentation dataset. You can use Kaggle, Roboflow, and GitHub for different datasets. I chose a small dataset for segmenting roads, it contains 250 images. The image number doesn’t change anything about the pipeline, every step is the same.

There are two classes, background and road class. You can see an example annotation from the above image. Copy your images and masks inside the data/imgs and data/masks folders in the cloned repository. Masks must be images, not JSON files or TXT files. There are two export options for semantic segmentation datasets in Roboflow, and if you choose the “Semantic Segmentation Masks” option, it will give the mask as an image.
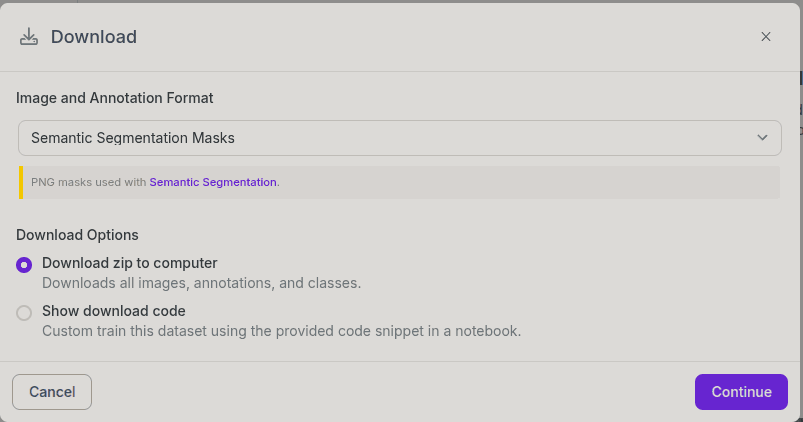
By the way, you can change these folder names (imgs and masks) inside the train.py file.
We will directly use the train.py file for training. Open a new terminal, activate your environment, and go to the cloned repository. Then, start training:
python train.py --epochs 12 --classes 2 --scale 0.5 --batch-size 1
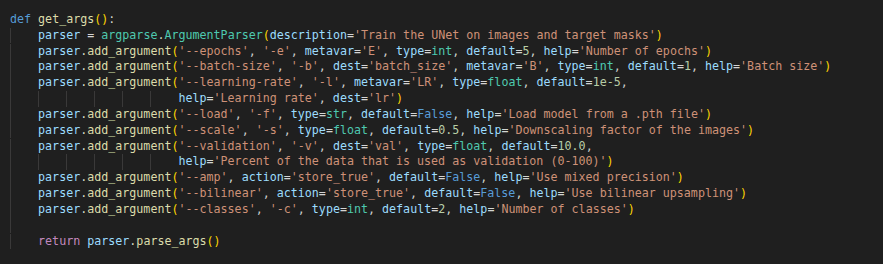
I have an old GPU (6 GB VRAM), and I got CUDA out-of-memory errors. So, I reduced the scale to 0.5. It reduces image sizes during training, therefore reducing memory requirements. You can increase or decrease the epoch number. If you used a different dataset, you should change the class number as well (class number + 1). Don’t forget to add one for the background. You can see all parameters in the below image:
Now, training has started, and it might take some time depending on your GPU, dataset, and configuration.
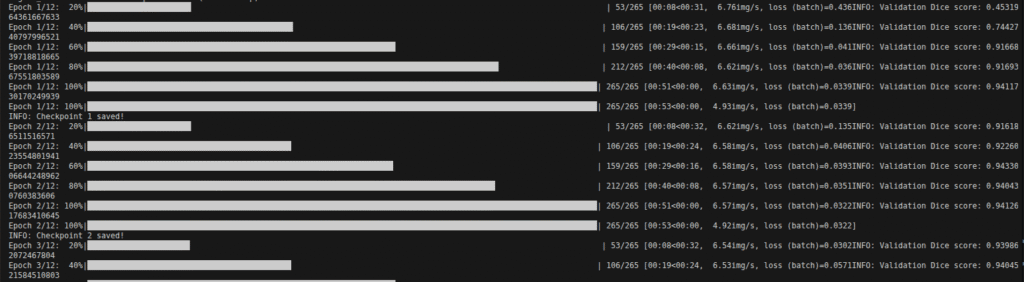
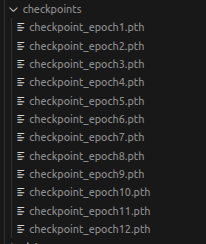
It automatically saves the model after each epoch inside the checpoints folder
You can directly use the predict.py file, or you can create a simple script and integrate the model into your pipelines, I will show you both.
Don’t forget to change path to the image.
python predict.py -i /images/test_image.jpg -o output.jpg --model checkpoints/checkpoint_epoch12.pth --classes 2 --viz


Now, let me share with you a simple script for inference. Let’s start with importing few libraries:
import torch
import matplotlib.pyplot as plt
from PIL import Image
import numpy as np
from unet import UNet
from utils.data_loading import BasicDataset
import torch.nn.functional as F
from IPython.display import display
Now, let’s load the trained model.
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = UNet(n_channels=3, n_classes=2)
state_dict = torch.load('checkpoints/checkpoint_epoch12.pth', map_location=device)
state_dict.pop('mask_values', None)
model.load_state_dict(state_dict)
model.to(device)
model.eval()
Let’s read and process the image.
# Load and preprocess image
img_path = "images/test_image.jpg"
img = Image.open(img_path)
img_array = np.array(img)
# Convert to tensor
img_tensor = torch.from_numpy(BasicDataset.preprocess(None, img, 1.0, is_mask=False))
img_tensor = img_tensor.unsqueeze(0).to(device=device, dtype=torch.float32)
Run the model:
# Run the model
with torch.no_grad():
output = model(img_tensor).cpu()
output = F.interpolate(output, (img.size[1], img.size[0]), mode='bilinear')
mask = output.argmax(dim=1).squeeze().numpy()
Display the result:
# Visualize prediction overlay
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(10, 10))
ax.imshow(img_array)
masked = np.ma.masked_where(mask == 0, mask) # Show only segmented regions
ax.imshow(masked, alpha=0.5, cmap='jet')
ax.axis('off')
plt.tight_layout()
plt.savefig('prediction_overlay.png', bbox_inches='tight', dpi=150)
plt.close()
# Display result
overlay_img = Image.open('prediction_overlay.png')
display(overlay_img)

That’s it from me, see you in another article, babaay 🙂